How to Structure an ML Project in the Era of LLM Assistants

I joined the Yale/UNC-CH - Geophysical Waveform Inversion Kaggle competition to test drive and improve my ML workflow. My primary objective was to integrate LLM coding tools at every stage of the workflow and to gain a better understanding of what these tools are capable of.
Below, I discuss:
- How to structure your code for fast iteration
- Jupyter Notebook techniques that allow you to:
- explore the problem
- develop functionality quickly
- and execute long training runs
- The experience of using LLMs for ML engineering work
- What I could have done better in this competition
We begin with a brief overview of the problem and then proceed to discuss the points mentioned above.
Problem overview
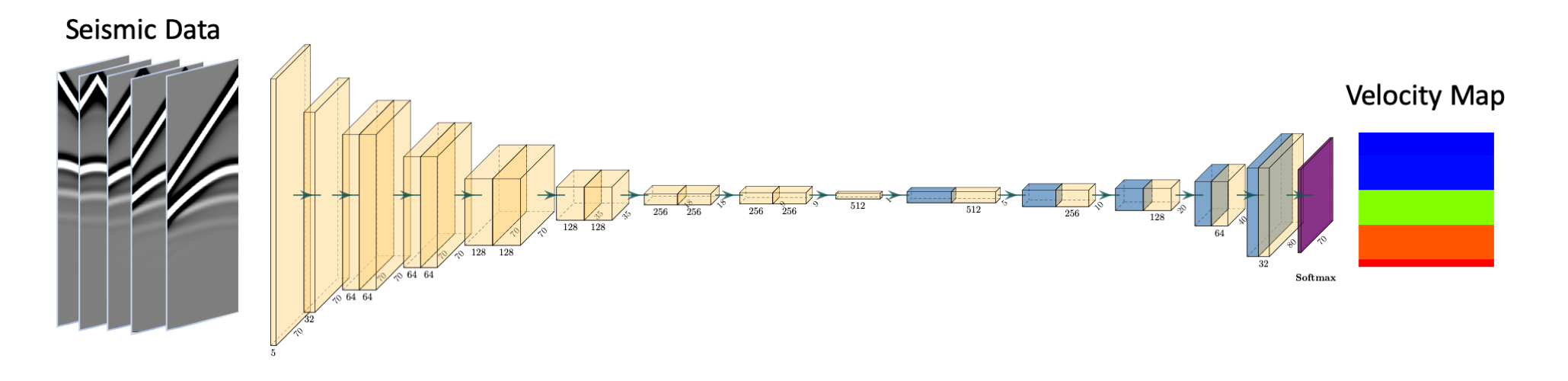
The competition was about creating velocity maps from seismic wavefield information.
At its core, the challenge can be summarised as going from a representation of dimensionality 5, 70, 1000 to a representation of dimensionality 70, 70.
The competition had a lot going for it:
- The problem was interesting and challenging
- There was a large amount of high-quality train data
- The train and test data were from very similar distributions
I joined the competition late, just 30 days before the finish. Ultimately, I placed 44th out of 1365 teams.
I ran all the experiments on GH200 on Lambda Labs, which is an absolute beast of a system. For working on remote machines (I had up to 5 GH200s spun up at a time), I followed the approach I shared here.
I share the refactored code that demonstrates the techniques discussed in this post as a GitHub repository. Running the code from this repository will give you a score of 19.19, which would have placed you in position 33 on the private leaderboard.
How to iterate quickly
There is only one thing that matters when beginning to work on an ML project – how quickly and how well you can learn about the problem space.
Whether it is a competition or a project at work, the same rules apply. In either scenario, you are expending valuable resources (most notably, your time) to make progress on solving a particular problem.
Kaggle competitors are masters at this game. This competition had a lot of great knowledge sharing, including Kaggle kernels from top competitors (such as the ones here or here).
When we study the kernels of people who consistently perform well, what do we see?
- A flat code structure that lends itself well to change and rapid prototyping
- Great understanding of the used libraries, leveraging advanced functionality with very few lines of code
The above is what I strove to incorporate into the reshaping of my ML workflow.
Here are some of the highlights of the techniques I adopted.
The flat structure
Why are Jupyter Notebooks so useful for machine learning?
Since machine learning is mostly about exploration and finding good solutions, you want to be able to scrutinize and modify each component of your pipeline with ease, both as you develop it and further down the road.
When working in a notebook, you can stop the execution at any point and inspect the pieces you created. This allows you to modify them or recombine them in novel ways.
Yes, I am fully aware of the Python debugger and your ability to halt execution at any point when using Python scripts.
Favorite recent jupyter notebook discovery - the %debug magic:
— Radek Osmulski 🇺🇦 (@radekosmulski) December 26, 2017
1. Get an exception.
2. Insert a new cell, type %debug and run it.
An interactive debugger will open bringing you to where the exception occurred and allowing you to look around! pic.twitter.com/9DSnSbpu15
A Twitter thread on combining the use of the debugger with Jupyter Notebooks
But the experience that Jupyter Notebooks offer in this regard, using the debugger or not, is unparalleled.
It is not so much about whether your tools make something possible, but about how easy and frictionless they make achieving the thing you are after.
So, if pausing execution and taking a look around at any point of our solution is the main benefit of using notebooks, why do we often write code in a way that limits our ability to take advantage of this?
What are the two ways in which we make inspecting and iterating on our code harder when working in Jupyter Notebooks?
The first obstacle that we introduce is wrapping our functionality in functions.
You first spend your time creating the function, and subsequently, whenever you want to explore new ways of doing something (or inspect our current functionality), you need to unwrap your code from the function or use the debugger.
Both approaches are tedious and error-prone. Additionally, the debugger in Visual Studio Code—my preferred environment for notebooks, particularly for its LLM integration—is difficult to use.
The solution is to minimize function creation. Since notebooks execute top to bottom in their finished form, start by writing code imperatively, like a recipe. Only extract functions when you have a compelling reason: when you genuinely need to reuse code elsewhere.
However, machine learning is fundamentally exploratory. Most of your code will change as you experiment, making premature abstraction counterproductive. The goal is to iterate quickly and probe the problem space for the actual shape of the problem with minimal friction.
Code complexity should never limit your experiments—whether by making them impossible or so unpleasant that you abandon the attempt.
Thus, a flat structure is the approach. Here is a relatively sophisticated training loop implemented directly in a Jupyter Notebook:

I can modify the training loop with very few keystrokes.
Plus, nearly all the information I need to understand how something works or to make a decision is contained in this cell or cells directly above it.
Imports that support flat code
You might have some code you don't expect to change and that you want to reuse across notebooks.
You could create a Python module (utils.py), but that makes it harder to inspect the code and iterate on it.
Nbimporter to the rescue! It gives you the ability to import from notebooks as you would from a Python module.
You can define your helper functionality in a notebook:
And then import it by first importing nbimporter and using the familiar import syntax:
However, we can take this pattern even further.
Sometimes, you might want to define the functionality you'd like to be importable, but you might also want to include diagnostic code or some other code useful for continuing development.
This is the case for me in the custom_unet.ipynbnotebook.
I first define the neural network architecture as a class. I create a cell with # STOP_WHEN_IMPORTED, and I proceed to instantiate the architecture in the notebook below it!
Now, when I run the notebook to make any modifications, I can immediately test them using the code below the # STOP_WHEN_IMPORTED statement
As a side note, the bit of functionality enabling this pattern is defined in notebook_import.py and has been one-shot by Claude Code.
Creating small utility functions like this using LLMs is a great use case!
Using Jupyter Notebooks for extensive experimentation
Jupyter Notebooks are for exploration and code development, but you do not want to run experiments in Jupyter Notebooks.
Any time you lose connection, you risk not being able to recover all the output.
Plus, how do you document the various experiments you ran? How do you run multiple experiments at the same time with different hyperparameters without modifying the code in your notebook?
The proposed solution is to use environment variables in combination with execnb by answer.ai.
Taking this approach, you can start a training run from the terminal (preferably, inside tmux) as follows:
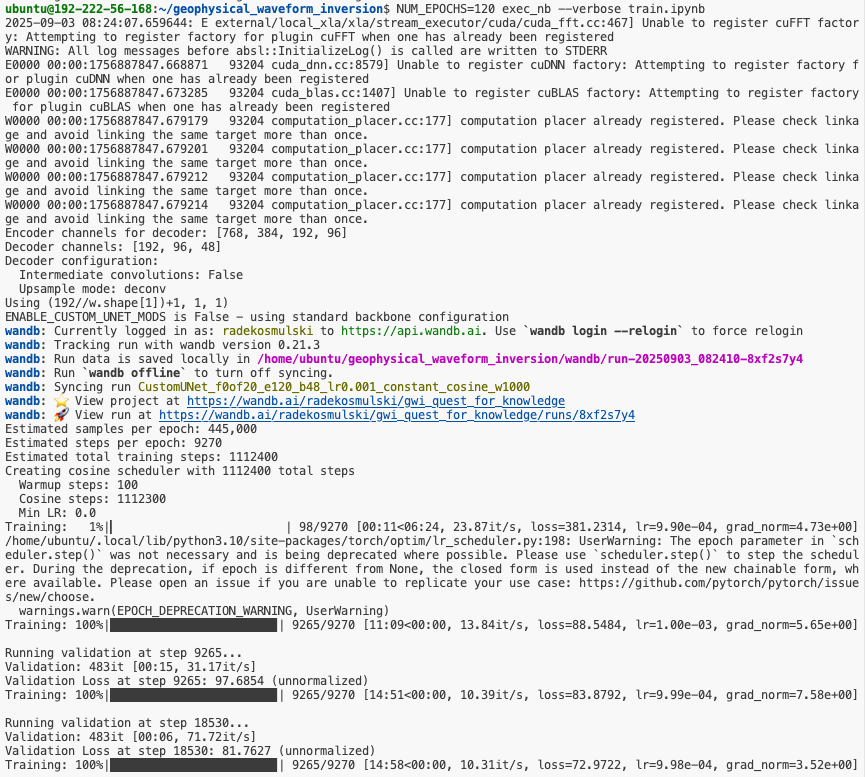
And what can you specify using environment variables? Anything you expose in your code!

train.ipynb exposes.There are risks associated with passing in hyperparameters in this way, one of them being that you might misspell a variable, and you won't get the effects you are after.
One solution is to add a print statement inside the notebook and visually confirm after triggering the run!
Is this ideal? No. Am I likely to explore other solutions? 100%.
Is this better than anything I have tried so far? Yes.
The point is, software engineering is full of trade-offs. I am willing to accept mild inconvenience (including a greater need to rely on my presence of mind while starting experiments, though I agree this is not ideal) so that I can optimize for what I am really after here – ease of change and speed of iteration.
Forking execution based on the runtime environment
As we rely on notebooks for development and experimentation, is there anything else we could do to make the experience even better for us?
Yes, there is!
You can conditionally run (or exclude from running) a subset of cells depending on whether you are running in a notebook or executing the notebook via execnb:
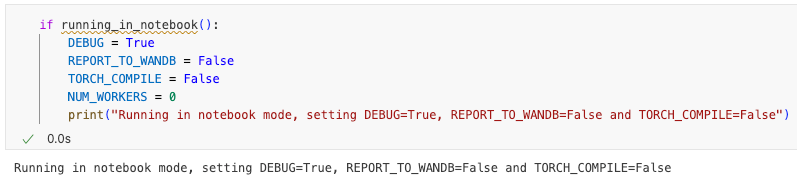
This functionality is defined in utils.ipynb and gives you unparalleled flexibility to further streamline and automate your workflow.
Again, it was one-shot by Claude Code.
Practical insights: LLMs in ML engineering
I bounce from thinking "LLMs are amazing, I want them to write all the code for me" to "I can't believe I spent so much time getting this code I got from an LLM to work !@#$%"
But I now believe that this is exactly the pattern you want to experience. Let me explain why.
Being on this sinusoid means two things:
- LLMs can be extremely powerful (how else would you arrive at being excited about their capabilities if not through experimentation and them doing something that impressed you?)
- Given your current understanding of their capabilities and how you use them, sometimes you end up in a lot of trouble. But as you continue to learn how to work with them, the valleys will become shallower and shallower, up to the point where you will nearly always stay above the 0 line.
This is precisely what I wanted to use this competition for. To put Claude Code to a serious test and learn in the process.
Despite moments of frustration, this collaboration has been extremely effective. It is an ongoing learning process; another leg in a journey I have been on for many months now.
Below, I share a couple of interactions with Claude Code and discuss its overall performance on this project.
The verdict: Was Claude Code useful?
For self-contained tasks, Claude Code with Opus as the driver is a beast.
For instance, in my refactored code that I am sharing, I wanted to see the impact of pseudo-labelling.

You can find the one-shotted script here. This is the level of responses you can expect from Claude Code:
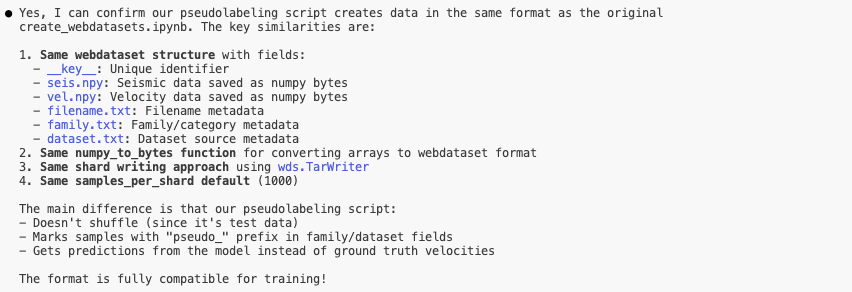
And yes, I was able to confirm that the script worked simply by predicting on the generated data and getting a loss close to 0.
A 280-line functioning script delivered in a matter of minutes!
How long would it have taken me to write this script myself? 15-minutes? Half an hour? Would I have enjoyed this work?
Would this have been the best use of my time?
On the other hand, machine learning is extremely unforgiving when it comes to subtle bugs. Being able to verify the correctness of code written by others and to write as bug-free code as possible yourself are very valuable skills.
In an earlier blog post on structuring machine learning projects, I wrote:
Test everything.
The only way to maintain your sanity in the long run is to be paranoid in the short run.
This entails extreme caution. The tendency of LLMs to hallucinate, or us not supplying the requisite context for a given task, can prove devastating.
The bottom line is that coding assistants don't fundamentally change how we solve problems with code.
They change how lines of code appear on our screens, but the structure of our solutions—and what dictates their quality—remains unchanged.
Putting Claude Code to the test
One way to improve performance in this competition was through data augmentation.
However, augmentation couldn't be done on the fly during training—creating augmented examples (beyond vertical flips) was computationally expensive, hence we had to apply the augmentations offline and store the newly created data to train on.
I delegated this entire process to Claude Code, providing only minor guidance on transformation parameters (like Gaussian noise levels) based on my intuition. Data generation ran for several days on my local workstation and required purchasing a 2TB HDD to handle the splitting and uploading to Kaggle.
This approach might seem counterintuitive: why spend significant computational resources on unverified outputs from LLM-generated code?
The answer lies in the type of resources used. While the process consumed hardware resources, it required minimal time and attention from me. The operation ran largely unattended, freeing me to focus elsewhere. Also, through iterative work with Claude Code, I had developed confidence in its ability to handle this specific task.
The results validated this approach. A baseline model without augmented data scored 29.56 on the private leaderboard. The same architecture and hyperparameters with augmented data achieved 26.96, a substantial improvement.
Regardless of the circumstances, the question is always the same: how to achieve more with less? I am 100% convinced that LLMs are a crucial part of the answer.
Reflecting on the competition – what went well and what could be improved
I spent a lot of time on two ideas:
- improving the architecture and testing / devising alternatives
- training on data represented as spectrograms
I tried other things along the way, but these were the two main themes.
I spent a lot of time on them, hoping a longer training run or yet another improvement to the architecture would send me up the leaderboard.
But the validation never really came. Yes, I created an okay solution. Placing 44th out of 1365 teams is no small feat, especially if you join a competition late and it is your first competition in years.
I feel I could have done so much more, though! Not in terms of the final placement—that was beyond my control.
But I could have done the problem better justice; I could have left fewer stones unturned.
Of course, I fell into the usual trap: overemphasizing architecture at the expense of working on data. But the core problem ran deeper.
How do you truly adapt the scientific method to ML?
How do you stay attuned to experimental results and validate ideas cheaply, rather than following preconceptions?
These are the questions I need to ponder.
But the overall approach to this competition worked wonders. I joined this competition with a specific goal in mind of what I wanted to practice.
And that worked great.
A truth I discovered in my life many moons ago is that project-based learning is extremely empowering and effective.
It is great to surf these waves yet again 🏄